PupDB
a database of pupylated proteins
PupDB
PupDB is a database of pupylated proteins and pupylation sites aiming to provide an easily accessible web service for the analysis of pupylated proteins. Hyperlinks to major protein, structure and annotation databases are provided for accessing related information. Four useful tools are constructed and integrated into PupDB to provide functions of browses, keyword searches, sequence similarity searches and interactive displays of protein structures.
[Back to content]
What information are stored in PupDB
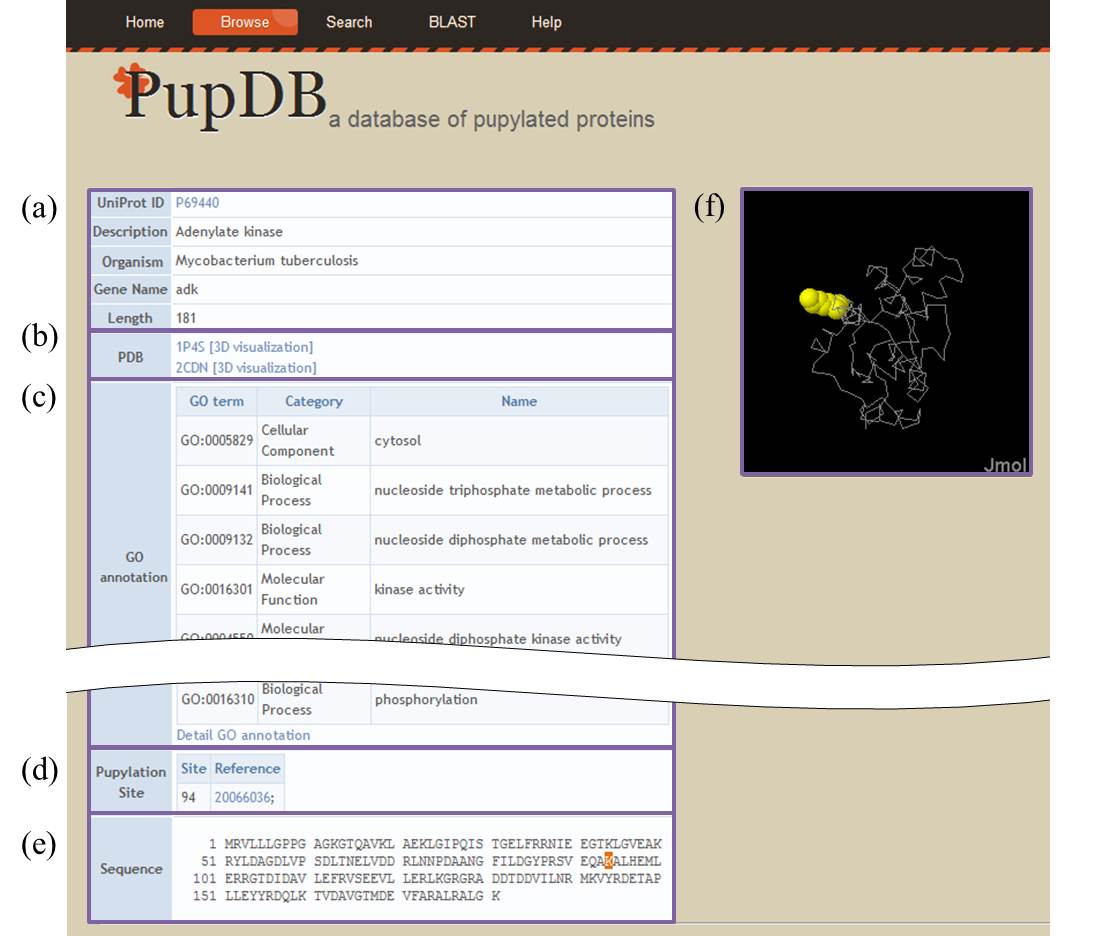
For each protein, the corresponding information consists of six major parts of basic information (a), PDB ID (b), gene ontology (GO) annotation (c), pupylation site (d), protein sequence (e) and structure (f).
Further GO information can be accessed by clicking the hyperlink of 'Detailed GO annotation' that links to the corresponding entry of QuickGO.
[Back to content]
How to Browse pupylatied proteins
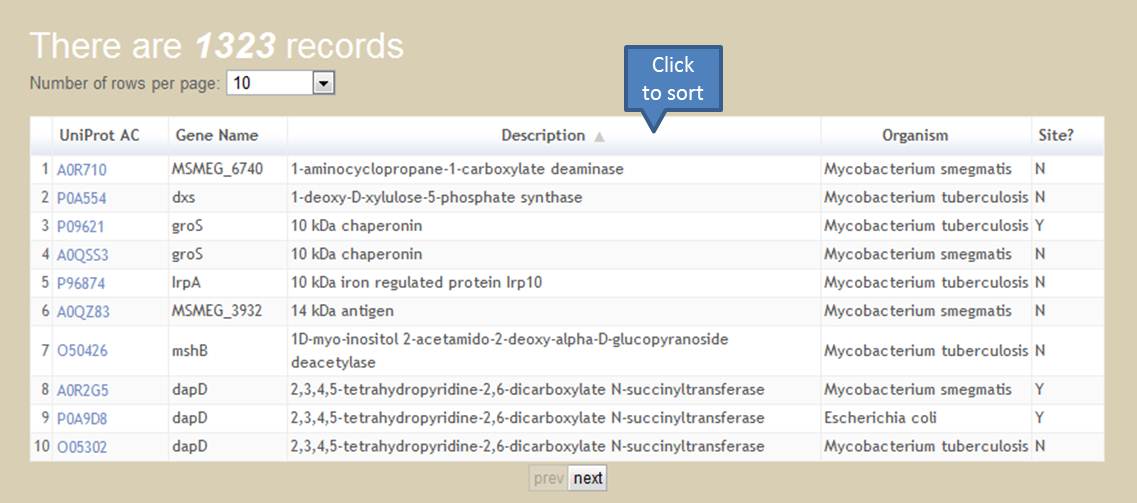
Users can browse all data collected in PupDB by selecting the 'Browse' option. All proteins will be shown in a sortable table. The entry with 'Y' in the field of 'Site' is a pupylated protein. Otherwise, it is a candidate pupylated protein with 'N' in the field of 'Site'. By clicking the caption of a specific column in a sortable table, the output table will be sorted according to data of the selected column. Furthermore, users can specify the number of rows shown per page.
[Back to content]
How to perform a keyword search
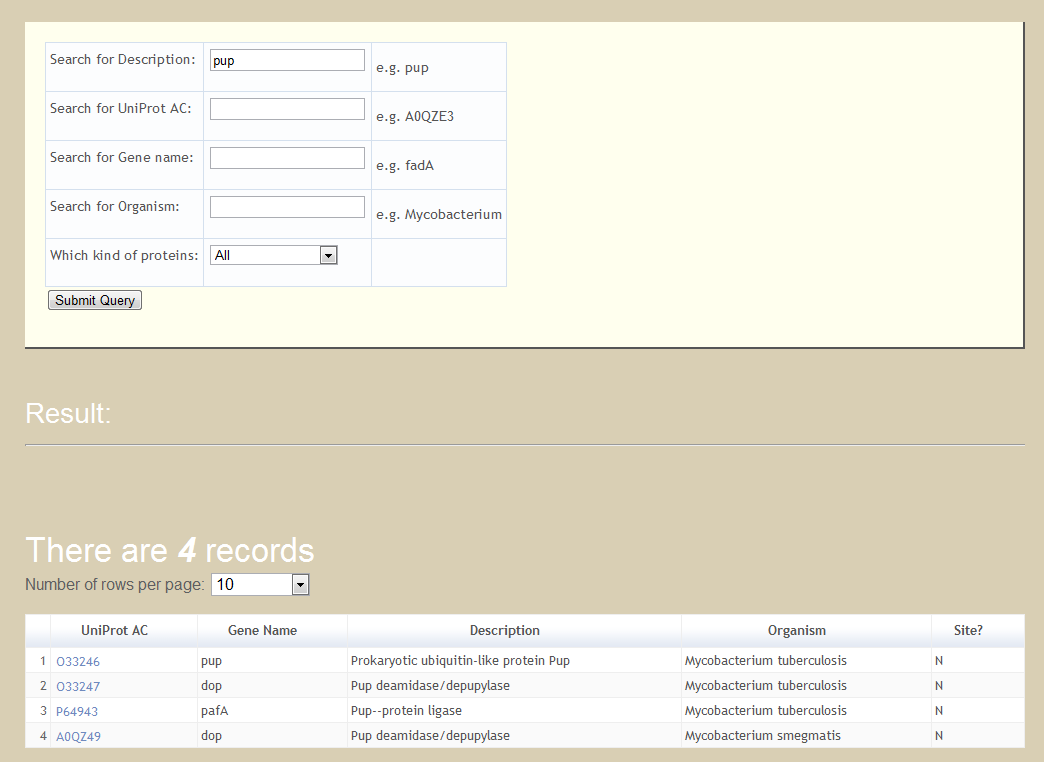
The tool of keyword search can be accessed by selecting the 'Search' option. There are five fields for searching PupDB including description, UniProt AC, gene name, organism and protein type. By entering keywords for any one or combination of the fields, PupDB will return search results as a sortable table according to the user input keywords.
[Back to content]
How to perform a BLAST search
Users can enter a protein sequence of interest in FASTA format to perform a BLAST [17] search against PupDB to fetch entries with a user-defined threshold of E-value. The BLAST tool can serve as a potentially useful tool for predicting promising pupylation sites by sequence similarity. In addition to the protein information, two additional columns of scores and E-values obtained from the BLAST search are included in the output sortable table. The detailed information of BLAST sequence alignment can be downloaded by clicking the download link.
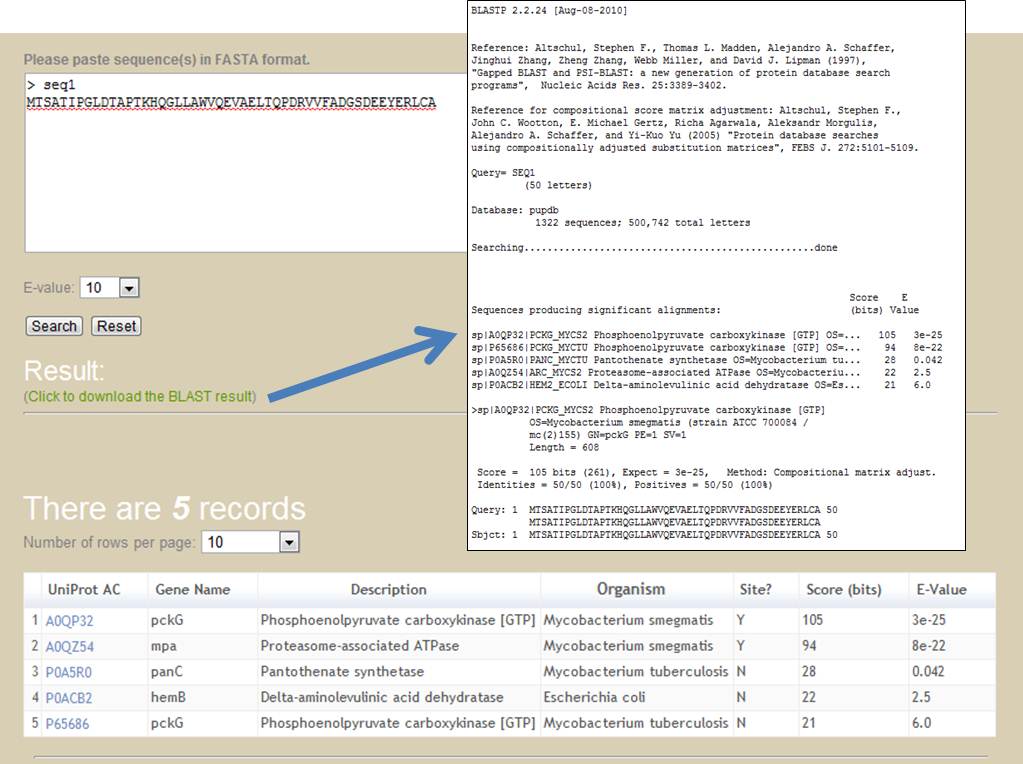
[Back to content]
How to download pupylated proteins
The exported tab-delimited table is available at http://cwtung.kmu.edu.tw/pupdb/pupdb_export.tab
[Back to content]
How to cite PupDB
If you find PupDB useful. Please cite:
Tung, C.-W. (2012) PupDB: a database of pupylated proteins, BMC Bioinformatics, 13, 40.
[Back to content]
References
- [Jmol] Jmol: an open-source Java viewer for chemical structures in 3D [https://www.jmol.org/ ]
- [BLAST] Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25(17):3389-3402.
- [PDB] Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res 2000, 28(1):235-242.
- [sortable table] Google Chart Tools [https://code.google.com/intl/zh-TW/apis/chart/index.html]
- [Pubmed] PubMed [https://www.ncbi.nlm.nih.gov/pubmed/]
- [UniProt] Magrane M, Consortium U: UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, 2011:bar009.
- [QuickGO] Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R: QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 2009, 25(22):3045-3046.
[Back to content]