iPUP
computational identification of pupylation sites
Content:
- iPUP
- How to submit a protein to iPUP web server for prediction of pupylation sites
- How to interpret the result of iPUP
- Download stand-alone software of iPUP for large-scale identification of pupylation sites
- How to use iPUP software for prediction of pupylation sites
- Datasets for download
- Prediction results for 1116 proteins in PupDB
- How to cite iPUP
- References
iPUP
iPUP based on the composition of k-spaced amino acid pairs and support vector machines is a tool for computational identification of pupylated proteins and pupylation sites. iPUP provides an easily accessible web service and a standalone software. The model of iPUP is trained on the dataset extracted from PupDB, a database of pupylated proteins and pupylation sites (link to PupDB).
[Back to content]
How to submit a protein to iPUP web server for prediction of pupylation sites
This is an example protein sequence of FASTA format
>P96382 MTFPGDTAVLVLAAGPGTRMRSDTPKVLHTLAGRSMLSHVLHAIAKLAPQRLIVVLGHDHQRIAPLVGELADTLGRTIDVALQDRPLGTGHAVLCGLSALPDD YAGNVVVTSGDTPLLDADTLADLIATHRAVSAAVTVLTTTLDDPFGYGRILRTQDHEVMAIVEQTDATPSQREIREVNAGVYAFDIAALRSALSRLSSNNAQQ ELYLTDVIAILRSDGQTVHASHVDDSALVAGVNNRVQLAELASELNRRVVAAHQLAGVTVVDPATTWIDVDVTIGRDTVIHPGTQLLGRTQIGGRCVVGPDTT LTDVAVGDGASVVRTHGSSSSIGDGAAVGPFTYLRPGTALGADGKLGAFVEVKNSTIGTGTKVPHLTYVGDADIGEYSNIGASSVFVNYDGTSKRRTTVGSHV RTGSDTMFVAPVTIGDGAYTGAGTVVREDVPPGALAVSAGPQRNIENWVQRKRPGSPAAQASKRASEMACQQPTQPPDADQTP
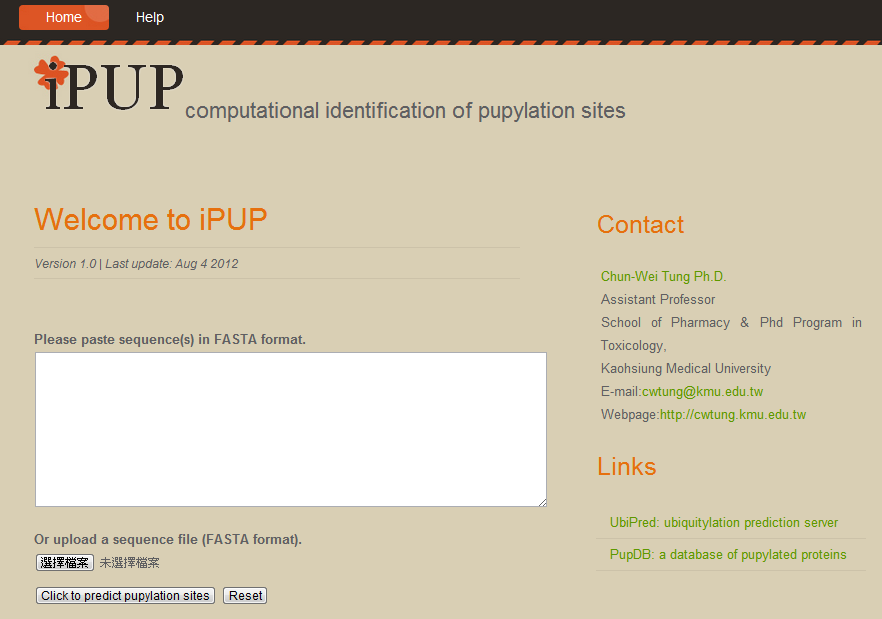
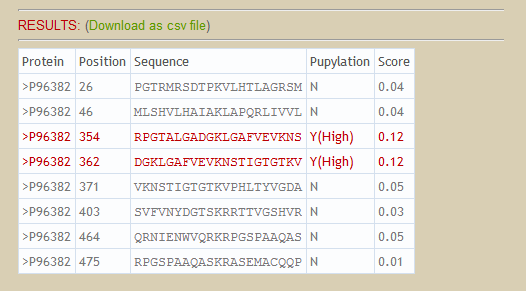
Also, there is a green link "Download as csv file". Users can download the prediction results for analysis.
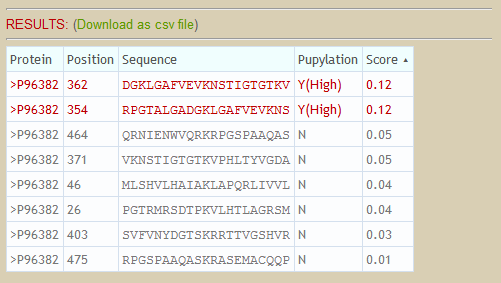
[Back to content]
How to interpret the result of iPUP
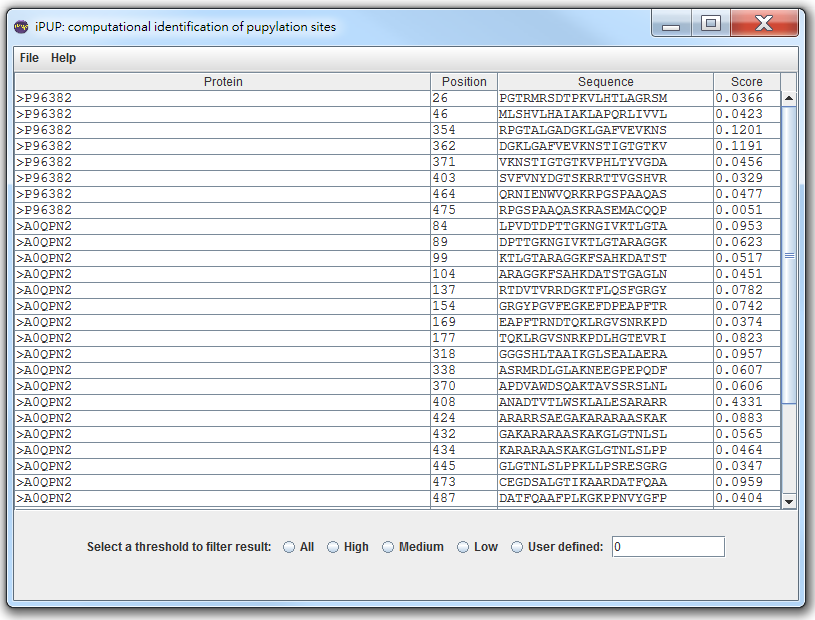
Given a protein sequence, the lysines (k) will be encoded as a modified composition of k-spaced amino acid pairs (MAAP). Subsequently, prediction results and probabilities will be shown in the web page. A set of pre-defined thresholds of High, Medium and Low is utilized to classify input lysines into four categories as follows. The thresholds are defined according to the specificity level of 10-fold cross-validation. Threshold values of High, Medium and Low are corresponding to specificity of 90%, 85% and 80%. Users can either use the pre-defined thresholds or define their own thresholds to classify lysines.
| Score | Probability of being a pupylation site |
|---|---|
| 0.1167 < score | Pupylation site(High) |
| 0.1044 < score < 0.1167 | Pupylation site(Medium) |
| 0.0963 < score < 0.1044 | Pupylation site(Low) |
| score < 0.0963 | Non-pupylation site |
[Back to content]
Download stand-alone software of iPUP for large-scale identification of pupylation sites
The JAVA-based software is available at http://cwtung.kmu.edu.tw/ipup/files/iPup.jar
[Back to content]
How to use iPUP software for prediction of pupylation sites
This is the content of an example file
Click to download example sequence file
>P96382 MTFPGDTAVLVLAAGPGTRMRSDTPKVLHTLAGRSMLSHVLHAIAKLAPQRLIVVLGHDHQRIAPLVGELADTLGRTIDVALQDRPLGTGHAVLCGLSALPDD YAGNVVVTSGDTPLLDADTLADLIATHRAVSAAVTVLTTTLDDPFGYGRILRTQDHEVMAIVEQTDATPSQREIREVNAGVYAFDIAALRSALSRLSSNNAQQ ELYLTDVIAILRSDGQTVHASHVDDSALVAGVNNRVQLAELASELNRRVVAAHQLAGVTVVDPATTWIDVDVTIGRDTVIHPGTQLLGRTQIGGRCVVGPDTT LTDVAVGDGASVVRTHGSSSSIGDGAAVGPFTYLRPGTALGADGKLGAFVEVKNSTIGTGTKVPHLTYVGDADIGEYSNIGASSVFVNYDGTSKRRTTVGSHV RTGSDTMFVAPVTIGDGAYTGAGTVVREDVPPGALAVSAGPQRNIENWVQRKRPGSPAAQASKRASEMACQQPTQPPDADQTP >A0QPN2 MSYTAADITELDDVQHTRLRPAVNLGLDVLNTALREIVDNAIEEVADPGHGGSTVTITLHADGSVSVADDGRGLPVDTDPTTGKNGIVKTLGTARAGGKF SAHKDATSTGAGLNGIGAAAAVFISARTDVTVRRDGKTFLQSFGRGYPGVFEGKEFDPEAPFTRNDTQKLRGVSNRKPDLHGTEVRILFDPAIAPDSTLD IGEVLLRAHAAARMSPGVHLVVVDEGWPGEEVPPAVLEPFSGPWGTDTLLDLMCTAAGTPLPEVRAVVEGRGEYTTGRGPTPFRWSLTAGPAEPATVAAF CNTVRTPGGGSHLTAAIKGLSEALAERASRMRDLGLAKNEEGPEPQDFAAVTALAVDTRAPDVAWDSQAKTAVSSRSLNLAMAPDVARSVTIWAANPANA DTVTLWSKLALESARARRSAEGAKARARAASKAKGLGTNLSLPPKLLPSRESGRGSGAELFLCEGDSALGTIKAARDATFQAAFPLKGKPPNVYGFPLNK ARAKDEFDAIERILGCGVRDHCDPELCRYDRILFASDADPDGGNINSSLISMFLDFYRPLVEAGMVYVTMPPLFVVKAGDERIYCQDESERDAAVAQLKA SSNRRVEVQRNKGLGEMDADDFWNTVLDPQRRTVIRVRPDESEKKLHHTLFGGPPEGRRTWMADVAARVDTSALDLT
In [File], you can choose [Open] to open the saved file.
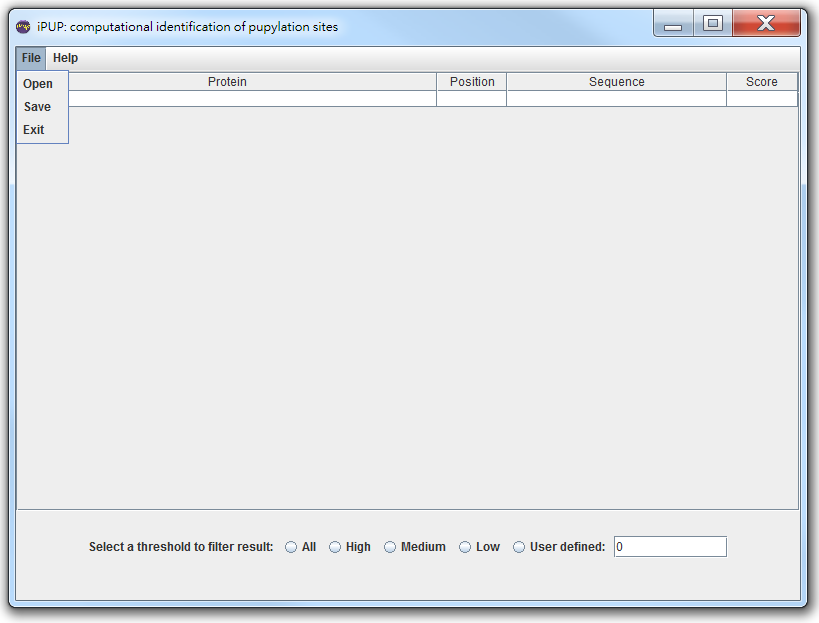
Or you can use [Help]->[Load sample Sequence]
Then, you can choose [All]/[High]/[Medium]/[Low]/[User defined] threshold to filter pupylation sites.
[Back to content]
How to cite iPUP
If you find iPUP useful. Please cite:
Tung, C.-W. (2013) Prediction of pupylation sites using the composition of k-spaced amino acid pairs, Journal of Theoretical Biology.
[Back to content]